Prof. Tingjian Chen’s Team Publishes a Paper in ACS Synthetic Biology——Development of Highly Stable Xenobiotic Nucleic Acid (XNA) Aptasensors for Bacillus cereus 5/B/6 Metallo-β-Lactamase
Recently, the research team led by Professor Tingjian Chen from the School of Biology and Biological Engineering, South China University of Technology, has made significant progress in the enzymatic synthesis of xenobiotic nucleic acids (XNAs) and their application in the selection of XNA aptamers, the construction of XNA aptasensors, and the biosensing of targets important in medicine and food safety. The team developed several highly stable and sensitive 2'-fluoro/methoxyl (2'-F/OMe)-modified RNA aptasensors for sensitive detection of Bacillus cereus 5/B/6 metallo-β-lactamase. This achievement has been published in the international journal ACS Synthetic Biology, entitled “Generation of stable 2'-F/OMe-RNA aptasensors for Bacillus cereus 5/B/6 metallo-β-lactamase”. PhD candidate Jing Wu, master's student Ziqian Xiao, and Associate Researcher Yuhui Du are co-first authors of the paper. Master's student Minglan Luo, PhD candidate Zhipeng Chen, and bachelor's student Binyuan Yang, Associate Researcher Pei Guo from the Hangzhou Institute of Medicine, Chinese Academy of Sciences, and master's student Jiacheng Zhang are co-authors. Professor Tingjian Chen is the sole corresponding author, and the School of Biology and Biological Engineering, South China University of Technology, is the first affiliation. (Link to the paper:https://pubs.acs.org/doi/10.1021/acssynbio.5c00547)
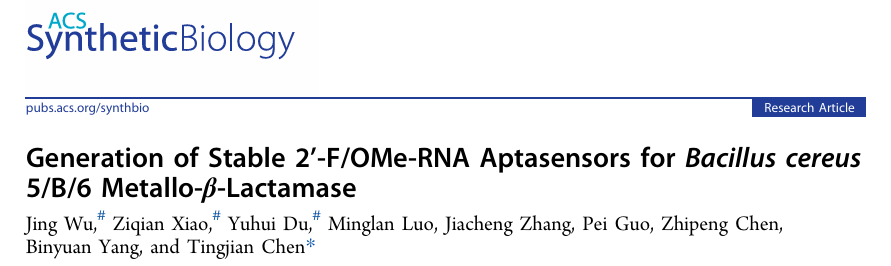
Aptasensors are widely used in biosensing and molecular diagnostics. However, conventional DNA/RNA aptamers are susceptible to nuclease degradation, limiting their application for target detection in complex biological samples. 2'-F and 2'-OMe-modified RNAs possess excellent nuclease resistance, making them ideal materials for constructing highly stable aptasensors. However, their efficient synthesis and library selection still face significant technical challenges.
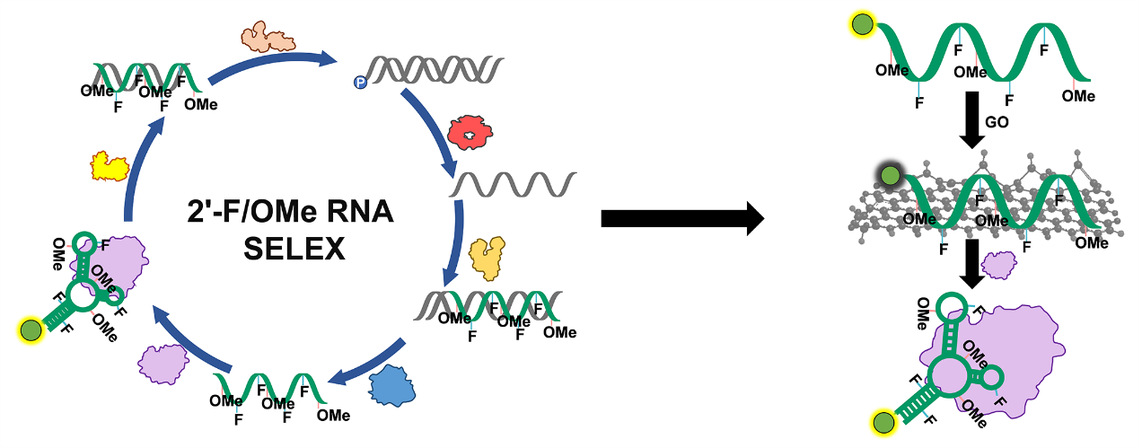
In this work, utilizing a laboratory-evolved xenobiotic nucleic acid polymerase, SFM4-6, the team efficiently synthesized a 2'-F/OMe-RNA library composed of 2'-OMe-A, 2'-F-U, 2'-F-C, and 2'-OMe-G nucleotides. Based on this, they established a complete systematic evolution of ligands by exponential enrichment (SELEX) system for selection of XNA aptamers, successfully identifying three high-affinity 2'-F/OMe-RNA aptamers (Apt-FUC1, Apt-FUC2, and Apt-FUC3) targeting B. cereus 5/B/6 metallo-β-lactamase. Surface plasmon resonance (SPR) analysis revealed that the dissociation constants for these three aptamers binding to the target protein were 95.9 nM, 70.0 nM, and 81.3 nM, respectively, demonstrating excellent target-binding capability. Further studies showed that the obtained 2'-F/OMe-RNA aptamers exhibited significantly higher biological stability in the presence of DNase I and in fetal bovine serum compared to their DNA counterparts. Building on the selected high-performance aptamers, the team further constructed three graphene oxide-based fluorescent 2'-F/OMe-RNA aptasensors. These aptasensors demonstrated extremely high sensitivity for detecting B. cereus 5/B/6 metallo-β-lactamase, with limits of detection (LOD) of 0.38 nM, 0.27 nM, and 0.29 nM, respectively. Even under condations containing DNase I, the aptasensors retained nearly 100% of their signal generation capability, significantly outperforming DNA aptasensors. Furthermore, these aptasensors exhibited good target detection specificity and stability in complex biological samples such as milk and 10% fetal bovine serum, accurately distinguishing the target protein from other β-lactamases and interferences.
This study establishes a complete technical pathway from the selection of XNA aptamers to the construction of XNA biosensors. It not only provides a new tool for the rapid detection of metallo-β-lactamases but also lays a methodological foundation for developing other highly stable XNA aptamers and aptasensors. It holds broad application prospects in fields such as biomedicine, food safety, clinical diagnostics, and environmental monitoring.
This work was supported by the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2019ZT08Y318), Guangdong Provincial Pearl River Talents Program (2019QN01Y228), National Key R&D Program of China (2019YFA0904102), National Natural Science Foundation of China (21978100), and Science and Technology Project of Guangzhou, China (2023A04J1470).