Neutrophils are the most abundant vertebrate leukocytes and they are essential to host defense. Dysregulation of neutrophil development is known to be associated with a variety of human diseases, such as severe congenital neutropenia, myelodysplasia syndrome (MDS), and leukemia [1]. Despite extensive investigation, the molecular network controlling neutrophil differentiation remains incompletely understood. Growth factor independence 1 (GFI1) is a transcriptional repressor predominantly involved in the formation of various blood lineages [2]. Dysregulation of GFI1 function often leads to the initiation and progression of various blood diseases. Humans bearing GFI1 mutations develop SCN [3], moreover, GFI1 mutations have been associated with acute myeloid leukemia (AML) and are a possible risk factor for myeloproliferative disorder with progression to AML [4]. Thus, clarifying the role of GFI1 in myelopoiesis and establishing the disease model will help in the treatment of GFI1-related diseases.
On August 10, 2021, Yiyue Zhang's group published their study titled Genetic and epigenetic orchestration of Gfi1aa-Lsd1-cebpa in zebrafish neutrophil development in Development. The study utilizes zebrafish to reveal Gfi1aa, Lsd1, and cebpa form a regulatory network that regulates neutrophil development and gfi1aa mutants develop an MDS-like phenotype, which will help to understand GFI1 related hematopoietic mechanism and disease pathogenesis.

The authors first generated the gfi1aa-deficient zebrafish mutant and found gfi1aa deficiency resulted in neutrophil lineage expansion. Then they classified the neutrophils and found gfi1aa deficiency leads to excessive neutrophil progenitors. Furthermore, by monitoring the proliferation rate of the neutrophil lineage, the authors found gfi1aa deficiency leads to excessive proliferation of neutrophil progenitors (Figure 1).
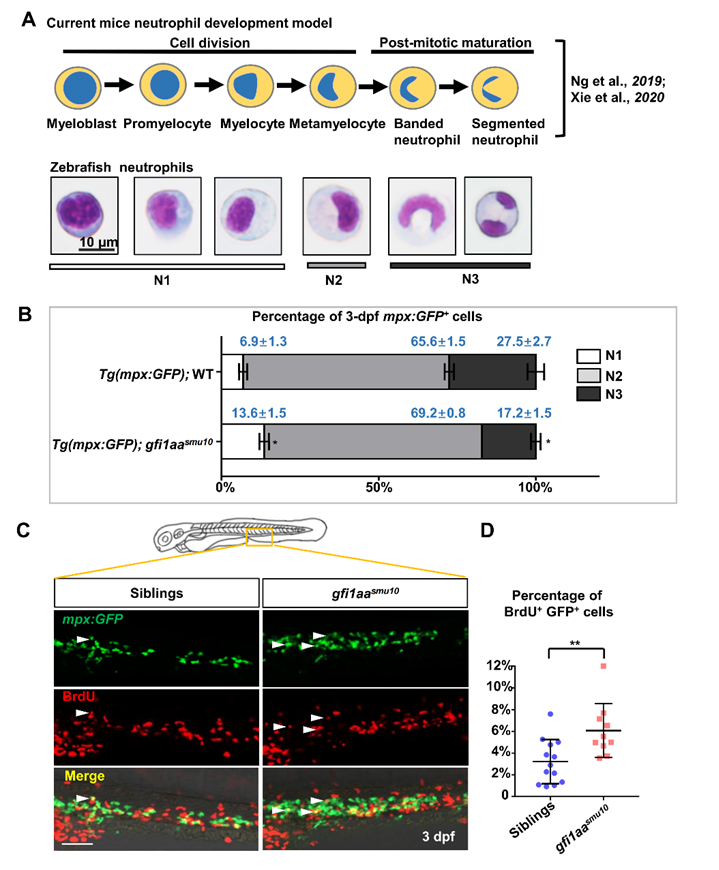
Figure 1. Proliferative neutrophil progenitors are accumulated in the gfi1aa mutant
As cebpa expression was increased in gfi1aa-deficient neutrophil and cebpa heterozygous could restore the number of neutrophil lineage cells in gfi1aa mutant to WT level, the authors performed luciferase reporter assay and found the cebpa transcriptional activity was significantly repressed by overexpression of Gfi1aa. Moreover, the authors performed Gfi1aa ChIP-seq and found Gfi1aa could bind to the transcriptional regulatory region of cebpa, suggesting Gfi1aa may repress cebpa expression by directly binding to its promoter region. Previous studies have shown that GFI1 could control hematopoietic cell fate by recruiting LSD1 [5]. The authors further demonstrated that Gfi1aa recruits Lsd1 to alter the cebpa H3K4 methylation status during granulopoiesis (Figure 2).

Figure 2. Gfi1aa epigenetically represses cebpa via Lsd1
As the neutrophil lineage proportion was abnormal during the embryonic stage, the authors monitored the neutrophil lineage proportion and blood phenotypes in adult gfi1aa mutants. They found with age, myeloid precursors in kidney marrow and neutrophils in peripheral blood were increased in gfi1aa mutants compared with WT, which resembles the hematopoietic phenotype of some MDS patients (Figure 3).
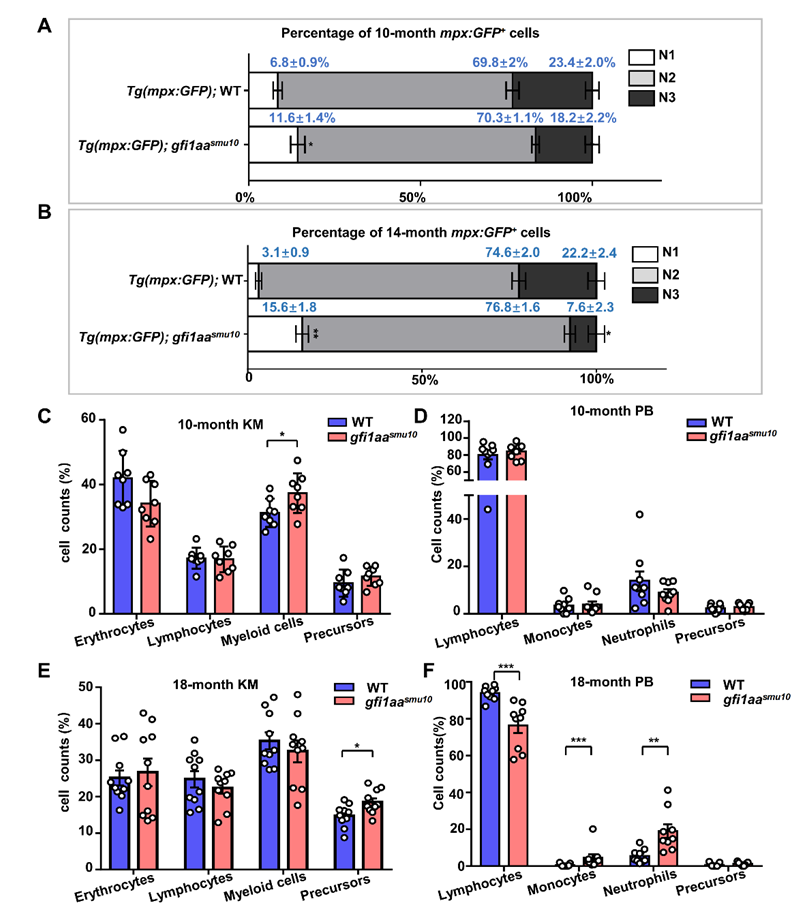
Figure 3. Adult gfi1aa mutants display hematological disorders
Taken together, the study reveals the role of the Gfi1aa-Lsd1-cebpa regulatory network in neutrophil development and finds gfi1aa mutants exhibit an MDS-like phenotype, which will be helpful in understanding and treatment of GFI1-associated myeloid disorders.
Dr. Mei Wu from South China University of Technology and Yue Xu, a master student from South China University of Technology, contributed equally to this work. Professor Yiyue Zhang from South China University of Technology is the corresponding author of the work. This work was supported by the National Key Research and Development Program of China (2018YFA0800200), the National Natural Science Foundation of China (31922023), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2019), and the Fundamental Research Funds for the Central Universities (2019ZD54).
Reference
1. Newburger, P. E. (2006).Disorders of neutrophil number and function. Hematology Am Soc Hematol Educ Program, 104-10.
2. Van Der Meer, L. T., Jansen, J.H. & Van Der Reijden, B. A. (2010). Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia, 24, 1834-43.
3. Person, R. E., Li, F. Q., Duan,Z., Benson, K. F., Wechsler, J., Papadaki, H. A., Eliopoulos, G., Kaufman, C.,Bertolone, S. J., Nakamoto, B., Papayannopoulou, T., Grimes, H. L. &Horwitz, M. (2003). Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet, 34, 308-12.
4. Khandanpour, C., Krongold, J.,Schutte, J., Bouwman, F., Vassen, L., Gaudreau, M. C., Chen, R., Calero-Nieto,F. J., Diamanti, E., Hannah, R., Meyer, S. E., Grimes, H. L., Van Der Reijden,B. A., Jansen, J. H., Patel, C. V., Peeters, J. K., Lowenberg, B., Duhrsen, U.,Gottgens, B. & Moroy, T. (2012). The human GFI136N variant induces epigenetic changes at the Hoxa9 locus and accelerates K-RAS driven myeloproliferative disorder in mice. Blood,120, 4006-17.
5. Velinder, M., Singer, J.,Bareyan, D., Meznarich, J., Tracy, C. M., Fulcher, J. M., Mcclellan, D.,Lucente, H., Franklin, S., Sharma, S. & Engel, M. E. (2016). GFI1 functions in transcriptional control and cell fate determination require SNAG domain methylation to recruit LSD1. Biochem J,473, 3355-69.

